REGIOCARD - Julien Barc

“REGIOCARD”
“Regulome and cardiac pathophysiology”
Biological and clinical markers available today avoid to propose an efficient preventive screening and early diagnosis of individuals at risk of sudden cardiac death (SCD). We count about 400 000 SCD per year in Europe with a large proportion of victims among individuals from the general population free from forewarning sign of SCD. Predict and prevent ventricular fibrillation (VF), encountered in 80-90% of the SCD, seems to be a central mechanism to decipher and to improve the prevention of the risk of fatal arrhythmia. During the past 20 years, with others groups, we participated studying rare cardiac disorders, considered as sensitized model for SCD, and identified genes and pathways for new targets and mechanisms for VF/SCD. However these studies focused mainly on rare variants (with large effect) located in the coding regions representing only 2% of the genome. This limited the dissemination and the applicability to the general population where molecular mechanisms and etiologies seem more complex.
The lack of relevant marker for sudden cardiac death (SCD) led us to investigate the non-coding region of the genome, seat of the gene regulation. We identified by genome wide association study on the Brugada syndrome (BrS), a rare cardiac disease at risk of SCD, 21 common variants increasing dramatically the risk to develop the disease and likely relevant to the broad problem of SCD. However converting the genetic findings to molecular mechanisms is challenging since signals identified are located within non-coding regions.
Hence, based on human cardiomyocytes, we will firstly characterize the human cardiac regulome by identifying the cardiac gene regulatory regions (GRRs) using ATAC-seq (Figure 1) and ChIP-seq approaches and the nature and location of the major cardiac transcription factors (TF) implicated in gene regulation. We will then specifically investigate the 21 BrS loci by identifying (1) the GRRs associated to the GWAS signals (2) the pathogenic variants and (3) the TF binding sites affected.
REGIOCARD aims also to decipher the pathophysiological consequences on the phenotype by performing chromatin conformation experiment to identify the gene target(s) of the GRR and perform a transcriptomic study to appreciate the impact on the gene target expression (Figure 2).
Collaborators: Dr Nathalie Gaborit (l’institut du thorax, Nantes), Dr. Jérémie Poschmann (CRTI, Nantes) Prof. Stephan Mundlos (Max-Planck-Institut, Berlin) and the Nantes GenoBiRD facilities.
Figure 1: ATAC-seq data representation from human cardiomyocytes with the identification of a GRR (Pink) where the known causal common variant rs6801957 is located. 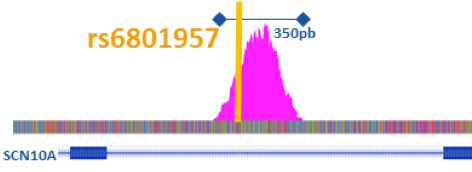
Figure 2: (Left) Schematic representation of the chromatin conformation between the SCN10A enhancer disrupted by rs6801957 and the SCN5A promoter.
(Right) Schematic representation of the impact on Nav1.5 expression.






